Bolster the growth and digital transformation of your business amid the outbreak through the Anti COVID-19 SME Enablement Program. Get a $300 coupon package for all new SME customers or a $500 coupon for paying customers.
By Hanchao from ApsaraDB
Genetic detection of pathogens is fundamental to the diagnosis of infectious diseases. The technique consists of five steps. (1) Collection of clinical specimens, such as venous blood samples, sputum, alveolar lavage fluid, or cerebrospinal fluid. (2) Culturing of the samples and extraction of nucleic acid from them. (3) Determination of nucleic acid sequences by using a high-throughput gene sequencer, which ensures accuracy by fragmenting long nucleotide sequences into small pieces between 50-200 bp in length. (4) Identification of and searching for matching sequences. (5) Analysis of the fragments to obtain their composition, that is, testing results. The testing results provide significant sources of diagnostic data and are used to formulate optimal treatment plans for individual patients.
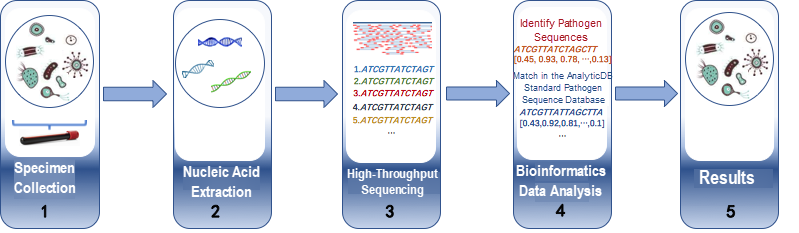
Figure 1. Pathogen detection process
For bioanalytical testing, performing pathogen detection usually generates about 500 million 75-bp gene fragments, of which some sample sequences are filtered. After the filtering, there will be about 100 million gene fragments to be queried. Normally, the nBlast[1] tool is used for comparing sequence information. This step can be very time-consuming for the whole detection process as it takes about 2 to 3 hours to complete. Alibaba Cloud AnalyticDB Vector Edition is an efficient gene sequence retrieval system, which completes the entire pathogen query and detection process in tens of minutes, effectively improving the performance of gene analysis.
Figure 1 shows the system interface for pathogen retrieval. In the following demonstration of our approach to pathogen retrieval, we included the base sequences of 12,182 viruses, segmented the viruses into 150-bp fragments (1,590,804 fragments in total), converted them into vectors, and stored vector data in AnalyticDB. In the search box, users can enter a gene sequence of interest and search the system database for identical or similar sequences. The gene sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), HIV, Ebola virus, and Middle East respiratory syndrome coronavirus (MERS-CoV) are used as samples to demonstrate the retrieval. Users can also copy similar gene sequences to test the query performance of the system.
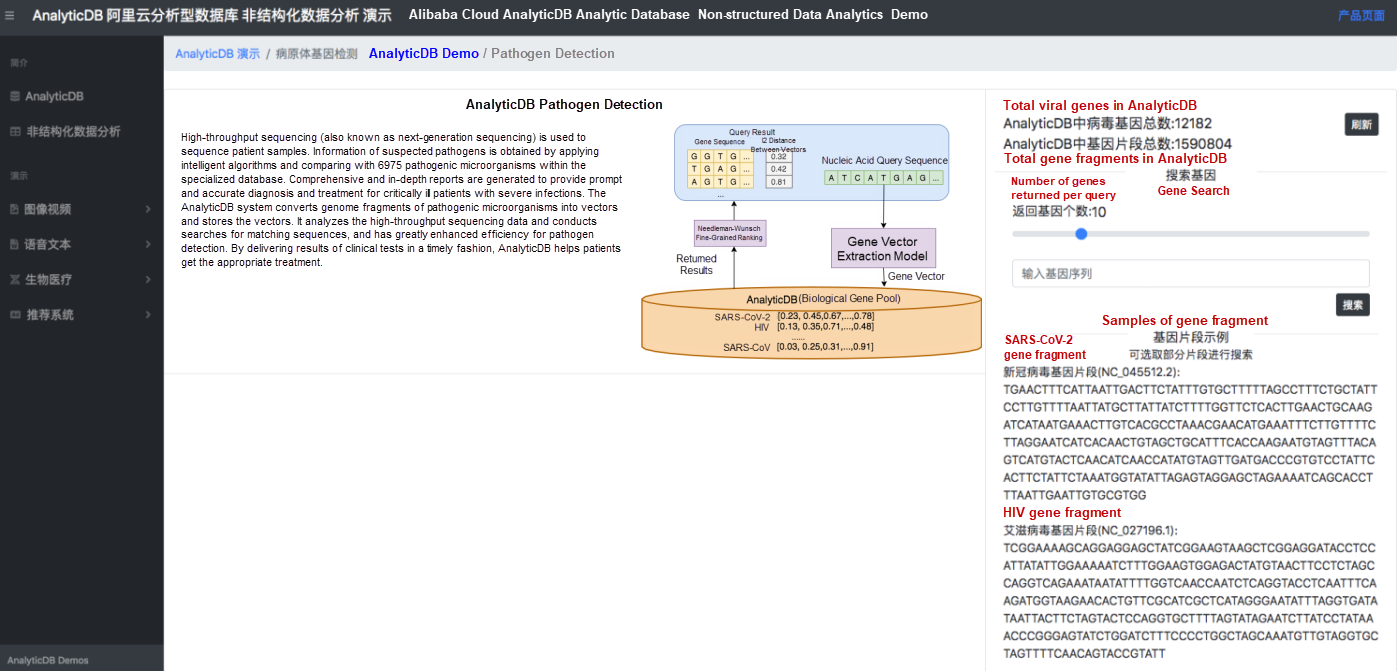
Figure 2. Demo of nucleic acid sequence query
As shown in Figure 3, the user entered a sequence segment of SARS-CoV-2 in the search box. As you can see, the SARS-CoV-2 sequence was listed first among all returned sequences. AnalyticDB currently provides an efficient method for vector similarity search, allowing the system to retrieve similar gene fragments in milliseconds.
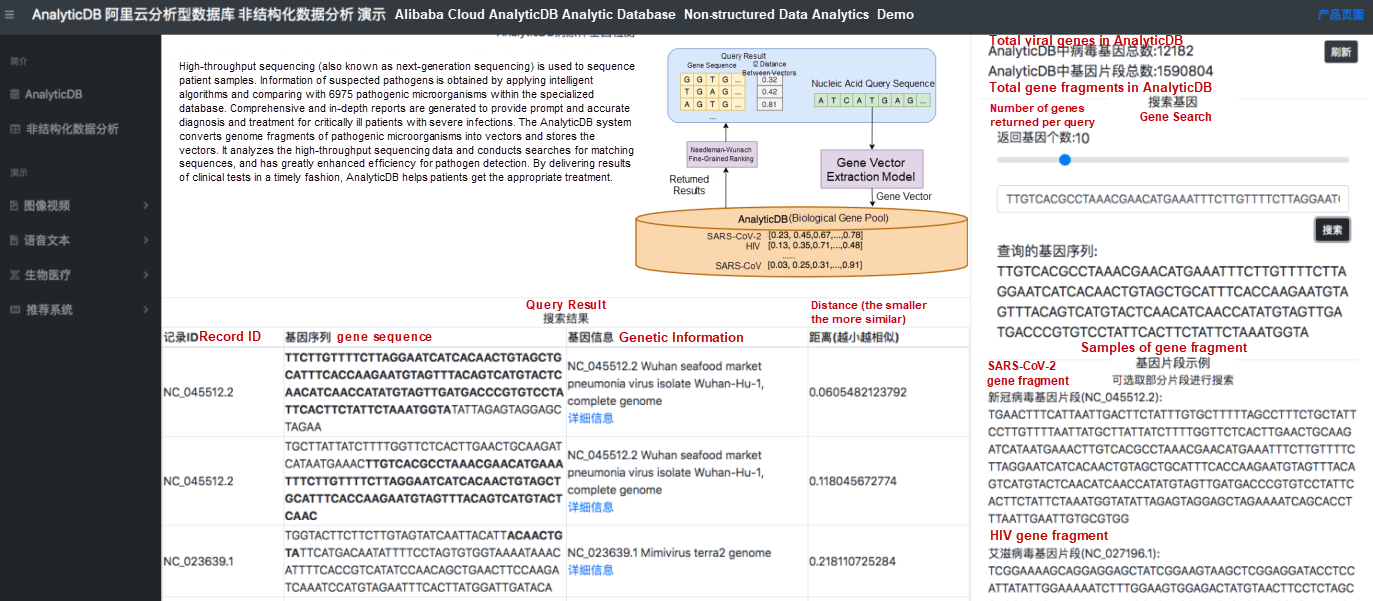
Figure 3. Retrieval results
By simulating human clinical samples and combining the strains of SARS-CoV-2 (accession number MT450872 [2]), SARS-CoV-2 (accession number MT450873 [3]), and MERS-CoV (accession number NC_019843.3 [10]), we can create a mixed sequence assembly of 75 bp for testing. This is to identify separate strains of SARS-CoV-2 and MERS-CoV from the strain mixture by searching against reference viruses in the database. Figure 4 shows the graphical display of matching results.
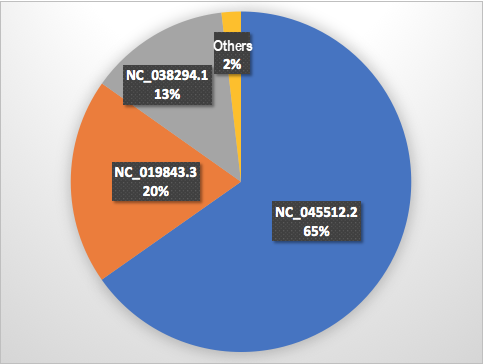
Figure 4. Matching genomes
The result showed that the system identified and returned three matching genomes. The accession numbers of these matches are NC_045512.2, NC_019843.3, and NC_038294.1. NC_045512.2 (65%) indicates the genome of SARS-CoV-2 (formerly called 'Wuhan seafood market pneumonia virus'), NC_019843.3 (20%) indicates the genome of MERS-CoV, and NC_038294.1 (13%) indicates the genome of betacoronavirus England 1, which is an isolate of MERS-Cov[8]. As per the analysis result in the preceding graph, the mixture contained SARS-CoV-2 and MERS-Cov.
The overall architecture of the Alibaba Cloud gene retrieval system is shown in Figure 5. In this architecture, AnalyticDB is responsible for processing all the application's structured data, such as gene sequence lengths, gene names, gene types, DNA and RNA information, and the storage and query of the feature vectors obtained from gene sequences. During query processing, we use a gene vector extraction model to convert genes into vectors and perform coarse-grained ranking in AnalyticDB. The classic Needleman-Wunsch[4] algorithm is used to rerank the result sets on a fine-grained level to return the most similar sequences.
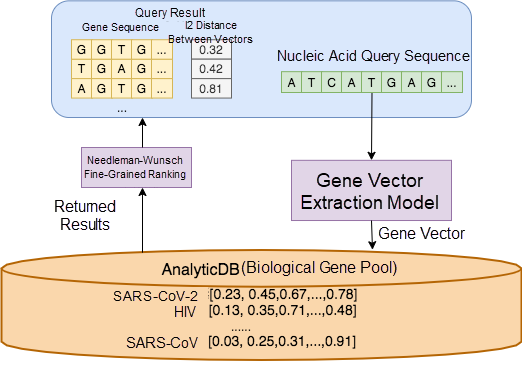
Figure 5. Gene retrieval framework
The training of the gene model has been explained in detail in the previous article[5]. We can obtain vectors of all k-mers by training the DNA k-mer model. As shown in Figure 6, five 8-mers are extracted from the 12-bp sequence. Next, we can convert the five 8-mers into corresponding vectors and then totalize and normalize the vectors to obtain the final vector of the 12-bp sequence. To further improve the accuracy of this approach, we can also apply other models like doc2vec[6] to learn the vector representation and convert complete sequences into vectors.
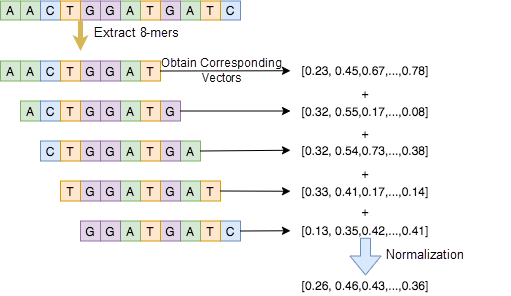
Figure 6. Convert DNA sequences into vectors
In this step, we trained two models. One was trained on all viruses and the other on 21 pathogenic bacteria (Propionibacterium acnes, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Escherichia coli, Acinetobacter baumannii, mycobacterium tuberculosis, Streptococcus pneumoniae, Klebsiella pneumoniae, Haemophilus influenzae, Haemophilus parainfluenzae, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Enterococcus faecium, Corynebacterium striatum, Human gammaherpesvirus 4 (Epstein-Barr virus), Torque teno virus, Human adenovirus B, Aspergillus flavus, Candida albicans, and Pneumocystis jirovecii). The genes were cut into 150-bp fragments, which were then converted into vectors and stored in AnalyticDB for retrieval. As a result, the data set contained 1,590,804 fragments of 12,182 viruses and 1,521,807 fragments of 275 genomes of the 21 bacteria.
Experiment 1 (see Table 1): One 75-bp fragment was randomly selected from the database. Since we knew this 75-bp fragment was extracted from which fragment and of which genome, we then searched for this 75-bp fragment in the database to check if the correct genome fragment would appear in the first n result sets. The precision of the top n retrieved result sets, or Precision(n), was calculated from the following formula. After calculations, we got the values (in percentage) shown in Table 1.
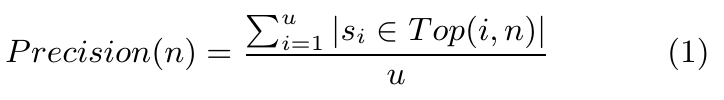
Here, n indicates the length of the list returned by the query, and u indicates the number of queries. In this experiment, u is 1,000. The expressionff |Si ∈ Top(i,n)| indicates whether sequence si is among the top-n returned items. It takes one of two possible values: "1" for yes and "0" for no. A smaller value of n indicates a higher precision and better retrieval performance of the system. As Table 1 shows, the precision values of top-20 ranked result sets were both over 99% for the two models, which qualifies our gene sequence retrieval system for use in identification in fragmented genome sequences.

Table 1. Precision values from genetic sequence detection
Experiment 2 (see Table 2): One 75-bp fragment was randomly selected from the database. We generated 2% random mutations in the sequence (mutation rates in the natural world are quite low, for example, each newborn baby carries roughly 30 mutations among 3 billion base pairs of the human genome. RNA viruses have relatively high mutation rates, but the rates are generally lower than 1%) and searched for this fragment in the database to check if the correct genome fragment would appear in the first n result sets. As the values in Table 2 show, although the precision were reduced after the mutation, the precision values of top-20 ranked result sets still reached 0.99.

Table 2. Precision values from mutated sequence detection
Experiment 3 (see Table 3): Comparison was made between the retrieval speeds of BLAST and the algorithm used in our system. A total of 9.7 GB of viral sequences, genome sequences of fungi, and some plant genome sequences[7] were downloaded and imported to the AnalyticDB database and the BLAST database. We performed 100 different queries and averaged the experimental results. On average, Blast took 3.22 seconds to return results for a query. Our algorithm took 0.257 seconds to complete a query (including the conversion of genes into vectors, coarse-grained ranking of vectors, and fine-grained ranking with the Needleman-Wunsch algorithm) with a precision of 0.95 among top 30 results, representing a 12.5-fold improvement over the BLAST speed.

Table 3. Retrieval time
[1] Blast+ https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/
[2] https://www.ncbi.nlm.nih.gov/nuccore/MT450872
[3] https://www.ncbi.nlm.nih.gov/nuccore/MT450873
[4] Needleman, Saul B. & Wunsch, Christian D. (1970). "A general method applicable to the search for similarities in the amino acid sequence of two proteins". Journal of Molecular Biology. 48 (3): 443-53. doi:10.1016/0022-2836(70)90057-4. PMID 5420325.
[5] Hanchao. Alibaba Cloud's Providing Efficient Gene Sequence Retrieval for COVID-19 Sequence Analysis https://developer.aliyun.com/article/753097?utm_content=g_1000111278
[6] Mikolov Tomas; et al. (2013). "Efficient Estimation of Word Representations in Vector Space". arXiv:1301.3781
[7] Genome data sets https://www.ncbi.nlm.nih.gov/genome/viruses/variation/help/flu-help-center/ftp/
[8] de Groot RJ Baker SC Baric RS et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013; 87: 7790-7792
[9] https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2
[10] https://www.ncbi.nlm.nih.gov/nuccore/NC_019843.3
[11] https://www.ncbi.nlm.nih.gov/nuccore/NC_038294.1
While continuing to wage war against the worldwide outbreak, Alibaba Cloud will play its part and will do all it can to help others in their battles with the coronavirus. Learn how we can support your business continuity at https://www.alibabacloud.com/campaign/fight-coronavirus-covid-19
How to Use the Smart Access Gateway (SAG) App to Secure Access Your Cross-border Cloud Resources

2,593 posts | 793 followers
FollowAlibaba Clouder - April 30, 2020
Alibaba Container Service - October 15, 2024
Alibaba Clouder - March 6, 2020
Alibaba Container Service - July 16, 2019
Alibaba Clouder - September 20, 2019
Alibaba Developer - September 7, 2020

2,593 posts | 793 followers
Follow Big Data Consulting for Data Technology Solution
Big Data Consulting for Data Technology Solution
Alibaba Cloud provides big data consulting services to help enterprises leverage advanced data technology.
Learn More Big Data Consulting Services for Retail Solution
Big Data Consulting Services for Retail Solution
Alibaba Cloud experts provide retailers with a lightweight and customized big data consulting service to help you assess your big data maturity and plan your big data journey.
Learn More ApsaraDB for HBase
ApsaraDB for HBase
ApsaraDB for HBase is a NoSQL database engine that is highly optimized and 100% compatible with the community edition of HBase.
Learn More Whole Genome Sequencing Analysis Solution
Whole Genome Sequencing Analysis Solution
This technology can accurately detect virus mutations and shorten the duration of genetic analysis of suspected cases from hours to just 30 minutes, greatly reducing the analysis time.
Learn MoreMore Posts by Alibaba Clouder