In order to win this inevitable battle and fight against COVID-19, we must work together and share our experiences around the world. Join us in the fight against the outbreak through the Global MediXchange for Combating COVID-19 (GMCC) program. Apply now at https://covid-19.alibabacloud.com/
By Sun Xiangzheng, nicknamed Yunshang at Alibaba.
As part of a global effort to combat the novel coronavirus disease (Covid-19), Alibaba Cloud provided relevant technologies in the construction of the Global Health Drug Discovery Institute Open Sharing Platform for the research to produce the drugs and treatments that will save countless lives. These technologies include high-performance computing (HPC)—the technology we will discuss today—as well as supercomputing clusters, and CPU and GPU hosts, cloud supercomputing, and artificial intelligence (AI).
Research institutions and universities can migrate their computing systems that are connected to drug and treatment development to Alibaba Cloud's Elastic High-performance Computing (E-HPC) solution. For this, Alibaba Cloud's relevant teams have worked around the clock to provide technical support and assistance along with follow-up services for these institutions.
In this article, we will be looking at just how E-HPC helps researchers in their work to process massive libraries concerning small molecules quickly in the drug candidate screening stages of research. We will also take a look at Alibaba Cloud's computing solution for the Global Health Drug Discovery Institute (GHDDI) and its open results sharing platform designed to help speed up research so that frontline medical teams can save more lives.
The drug development cycle is very lengthy, typically taking at least 10 years from the initial stages of research to when a drug is put on the market and made available to the public.
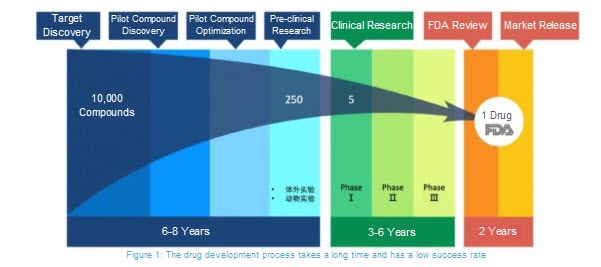
However, when responding to an epidemic, time is of the essence, as the health and security of the world is at risk. Therefore, scientists need to discover drug therapies quickly in order to save lives. One way to do this is by reviewing existing drugs that show promise as treatments for an epidemic disease. By using existing drug therapies, scientist can omit several of the steps typically involved with developing new drugs.
In the past, to discover promising compounds, researchers would need to conduct a large number of experiments. However, by using machines to simulate the interaction between molecular compounds and targets, scientists can quickly pinpoint potentially effective compounds, speeding up the process of finding promising leads faster.
High-performance computing or HPC, often also referred to as supercomputing, is an important tool for this kind of research. Moreover, the rise of cloud computing and high-performance computing, among other technologies and advancements, has in many ways fundamentally changed the way that scientists obtain computing power and use supercomputing services. As an example of this, Alibaba Cloud's E-HPC cloud supercomputing product allows scientists to build their own high-performance cluster systems in the cloud, allowing them to be able to build their desired computing platforms quickly and easily.
A huge amount of computing power is available in the cloud, and this computing power can be allocated flexibly. Scientists can purchase computing resources on demand, so they no longer have to worry about limited computing power as being a possible obstacle in their research for a new drug therapy.
However, you still may be wondering, how exactly does HPC related to viruses and drug research? To answer this question, we'll be taking a brief look into how viruses replicate and spread in hosts, how drugs can be used to inhibit the replicative capabilities of viruses, and finally we will discuss the role of HPC in drug research and development.
A virus consists of non-functional nucleic acid molecules, specifically DNA or RNA, which is surrounded by a protein coating. For an illustration of this, consider the diagram below of the tobacco mosaic virus, which is a virus that commonly infects the tobacco plant.
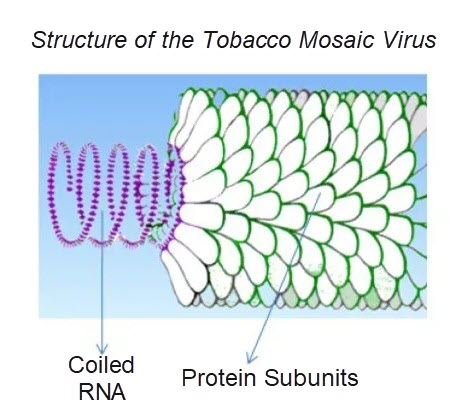
Viral populations cannot grow through cell division because they are acellular compounds. Instead, they use the machinery and metabolisms of a host cell to replicate.
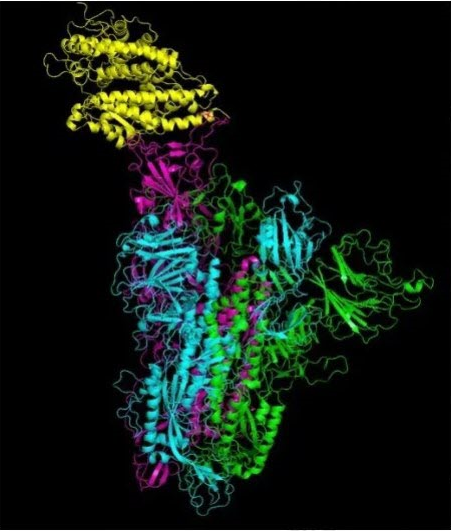
Coronaviruses, in particular, are a family of positive-sense single-stranded RNA viruses that have a lipid envelope studded with club-shaped projections. This family of viruses can cause diseases of varying severity in the respiratory tract, intestinal tract, liver, and nervous systems. Before the novel coronavirus, formally named SARS-CoV-2 , was discovered, two other types of coronaviruses were identified in the past twelve years, Severe Acute Respiratory Syndrome (SARS-CoV) and Middle East Respiratory Syndrome (MERS-CoV). Below is an illustration of the novel coronavirus.
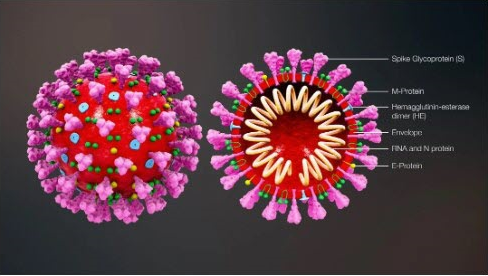
Now, let's discuss the viral replication process in detail. After a virus particle, or virion, enters a host cell, from a virus, the virus RNA is replicated and the virus protein is synthesized. Then, the RNA and protein are assembled into multiple new virions. Pictured below is an illustration of the molecular structure of viral protein.
A simplified diagram of the virus replication process is also shown below.
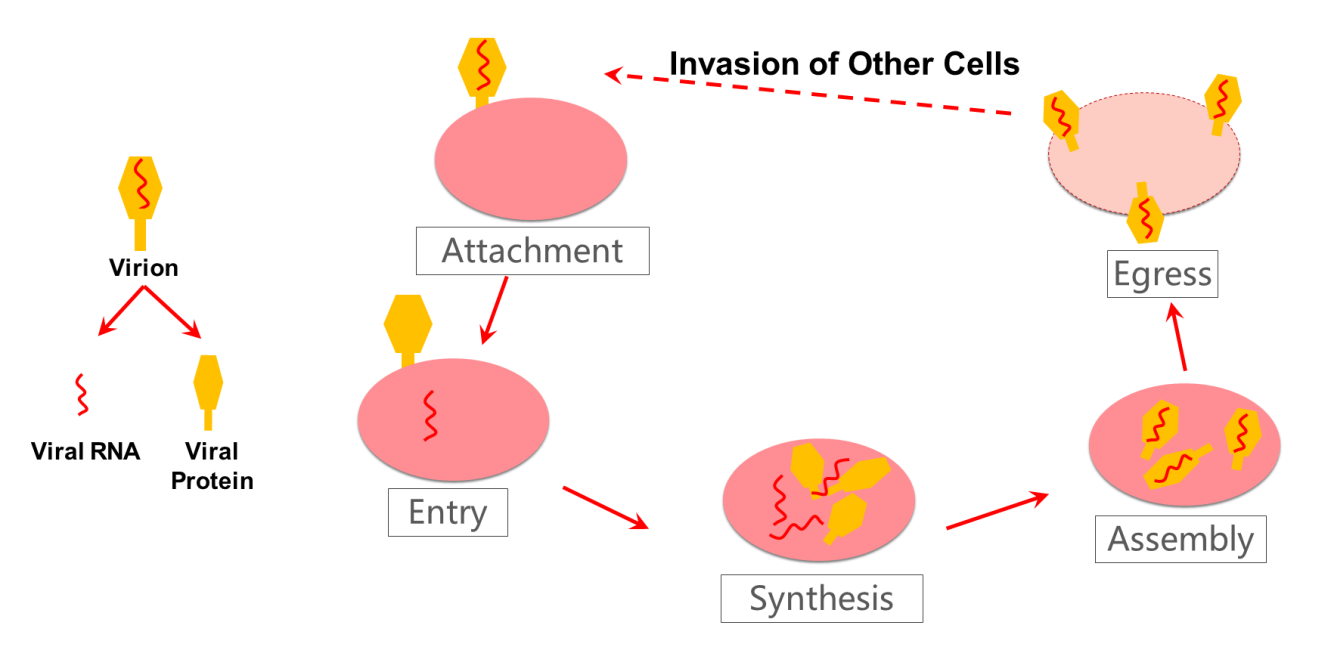
Drugs can be used to interfere with, or rather inhibit, the virus replication process to effectively prevent damage to the body. For example, in the synthesis process of viral proteins, protease intervention, such as 3CL protease and ProPL protease, can suppress the protease function. This is one method of virus inhibition. Other substances, such as ligand or drug treatments, can identify or bind with the protease. This process is called biological targeting. A ligand, which is a small molecule drug, can bind with the target of the viral protease, which in turn changes the three-dimensional structure of the protease, changing its function and inhibiting viral protein synthesis, thus preventing virus replication.
Drug development is a very complex and time-consuming process, and drug candidate screening is only one of the early stages in this lengthy process. In the search for small molecules that can bind to the viral protease structure, there is a large number of ligand libraries created by different research institutions, which contain massive amounts of data. Each library contains thousands of ligands. It is impossible to test them one by one in a timely manner. However, through computer value simulation, we can score the binding performance of different ligands and test the most promising ligands as candidate drugs, effectively accelerating the drug development process.
Due to the massive data volumes of ligand libraries, it is extremely difficult to filter promising ligands in a limited amount of time. For instance, if a ligand library has 10,000 candidate ligands and the average processing time is 1.5 hours for each candidate, it would take around 15,000 hours (or 625 days, almost 2 years' time) to process the entire library. Therefore, to complete the calculations within the specified time, the following conditions must be met:
To ensure efficient and smooth candidate screening, some computing services are also required, such as:
In addition to a computing platform, relative high-performance application software is required for drug candidate screening. Simulated drug candidate screening computation includes docking and molecular dynamics computation. Docking takes a relatively small amount of time and is often used for the preliminary screening of a large number of ligands. Popular docking software includes DOCK6, Autodock Vina, and Glide. Molecular dynamics simulation computing is time-consuming, and the time a test takes can vary. This process is used to further analyze the potential candidates selected in the docking process. Popular molecular dynamics software programs include Gromacs, Namd, and Amber. In this process, general-purpose GPUs (GPCGPUs) can provide a significant level of acceleration.
Cloud computing can serve as a means for drug research institutions to obtain the computing resources and services that are needed to build powerful high-performance clusters. As a part of tis solution, Alibaba Cloud provides several products and services, which are powerful tools for drug research and development. Besides Elastic High Performance Computing (E-HPC), Alibaba Cloud also offers cluster-based file sharing systems such as Network-Attached Storage (NAS) and Cloud Parallel File System (CPFS), along with several database management systems, which can be used in combination with E-HPC. Alibaba Cloud E-HPC on its own can allow users to quickly develop their own high-performance cluster systems in the cloud. Then, the other products can be used to configure the high-performance servers and large-capacity storage to be connected to these systems. This product also provides multi-node software runtime and high-throughput task processing solutions. In other words, E-HPC is ready to meet the computing platform requirements of drug researchers and developers across the globe.
The Alibaba Cloud E-HPC cloud supercomputing product is a cloud-native high-performance computing cluster solution. E-HPC integrates multiple Alibaba Cloud products, including:
E-HPC configures HPC job management and account management and integrates common HPC applications to provide users with a webpage-based operation interface. This allows users to obtain their own HPC clusters, hold root permissions, and manage and configure their clusters.
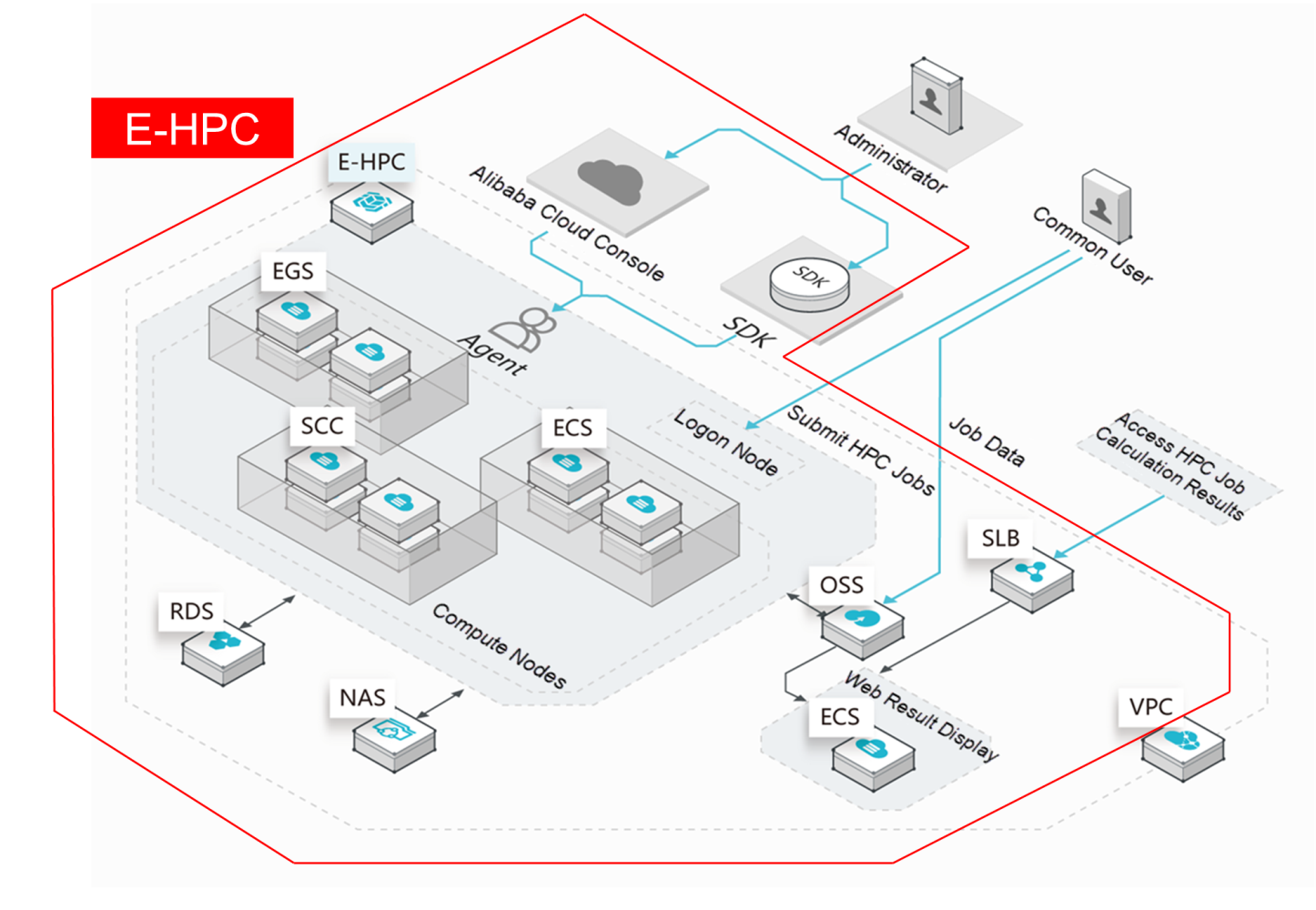
In addition to these functions, Alibaba Cloud provides a variety of computing instance types with diverse computing capabilities (including 1vCPU, 2vCPU, 4vCPU, up to 104vCPU), different memory ratios (including 1vCPU:2 GB, 1vCPU:4 GB, and 1vCPU:8 GB), and optional GPU or FPGA accelerator cards. Most of our CPU models use Intel's latest CPU architecture. Entirely developed by Alibaba Cloud, ECS Bare Metal instances serve as a new type of computing server product that is based on next-generation virtualization technology. ECS Bare Metal instances provide the elasticity of virtual machines and the performance and functionality of physical machines, putting the computing performance of the entire machine at the user's service. An ECS Bare Metal instance is equipped with a RoCE high-speed network that supports remote direct memory access (RMDA). This makes it a supercomputing cluster product, which can be used in large-scale and high-concurrency scenarios.
The HPC environment provides researchers with the computing platform they will need for drug research. However, to achieve efficient drug candidate screening, a high-throughput task solution is also required.
In the case, for instance, that we use DOCK6 to process a ligand library, a folder, such as mol2, stores a large number of small molecule files. The processing procedure for each small molecule file is the same, and all the files need to be calculated with the same receptor, such as a viral protease.
The code for a serial processing method is shown in the following figure. Here, dock.in is the file input using the DOCK6 command, and the corresponding parameter values need to be modified according to the small molecule file name. This code snippet traverses each molecule file in the mol2 folder, generating an input file named dock.in for each file, and then runs the DOCK6 command.
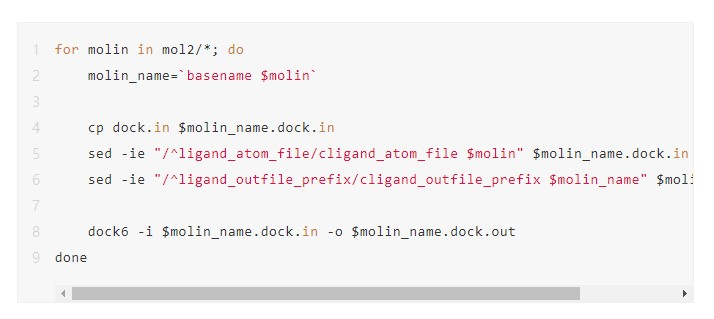
Serial execution takes a long time and cannot utilize the computing power of high-performance clusters. Therefore, we need to find a way to speed up the process by using multiple nodes and CPU cores of a cluster. This can be done in a variety of ways. For example, we can manually divide the mol2 folder into several subfolders, each of which contains a few small molecule files. Then, we can execute the files in each subfolder serially. This method requires a large amount of manual intervention. In particular, if a task encounters an error and needs to be re-submitted, this can easily lead to repeated or omitted computations.
E-HPC provides a high-throughput task solution. In this example, a large number of small molecule files can be concurrently processed in three steps.
1. Save the names of the molecule files under the mol2 folder to a text file, such as molin.

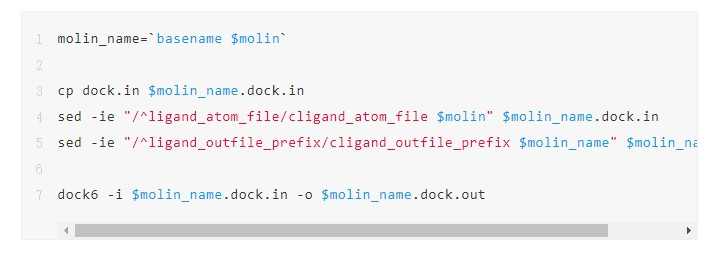
2. Develop a script named task.sh for processing individual small molecule files and replace the small molecule file name with $molin. Unlike the serial logic, you can see that the processing code is directly copied from the for loop.
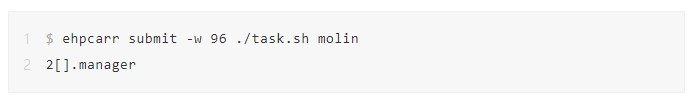
3. Run the E-HPC high-throughput task processing command ehpcarr to execute task.sh. The job number 2[].manager is returned. Now, the tasks are concurrently processed by using 96 CPU cores. If the node contains less than 96 CPU cores, the tasks are automatically assigned to multiple nodes. For example, when you use instances with 12 CPU cores, all molecule processing tasks run on 8 nodes.
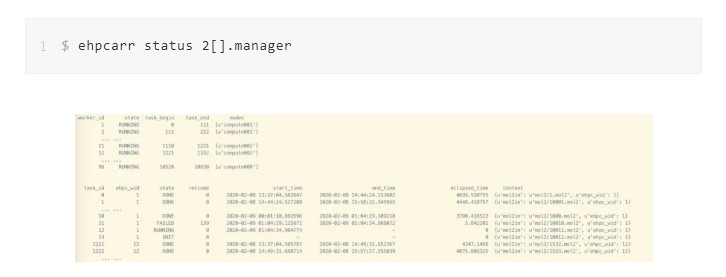
Run the ehpcarr command to query the concurrent execution status of tasks by job number. From the query results, you can obtain the current processing status of each task and its start and end times. Possible task states are DONE, RUNNING, FAILED, and INIT in queue. You can estimate the computing resources to be used for the next task execution based on the task execution duration. The query results show that:
The E-HPC solution is an array job solution based on the high-performance cluster job scheduler. It provides the following enhancements:
In the face of the current epidemic, sharing resources and research results is necessary as it can greatly accelerate the research progress and avoid repeated work. Understanding this, we helped to establish the Global Health Drug Discovery Institute (GHDDI).
The Bill and Melinda Gates Foundation, together with Tsinghua University and the Beijing municipal government jointly established GHDDI as an independent and non-profit drug research and development organization, and open sharing platform for drug research and development. The open sharing platform of GHDDI was built on Alibaba Cloud, developed wih E-HPC and build high-performance computing clusters for simulation computing in drug development. In addition, it creates different supercomputing sub-accounts for partners to allow the sharing of research results and computing resources.
At the same time, to facilitate the sharing and publishing of computing results on the E-HPC supercomputing cluster, storage using Alibaba Cloud Object Storage Service (OSS) is directly attached to the E-HPC supercomputing cluster. This means that the results are stored in OSS instances in the cloud. In addition, an ECS computing instance is created in the cloud to build a web server, and OSS access links are listed on the web server, allowing users to easily browse and download the stored results.
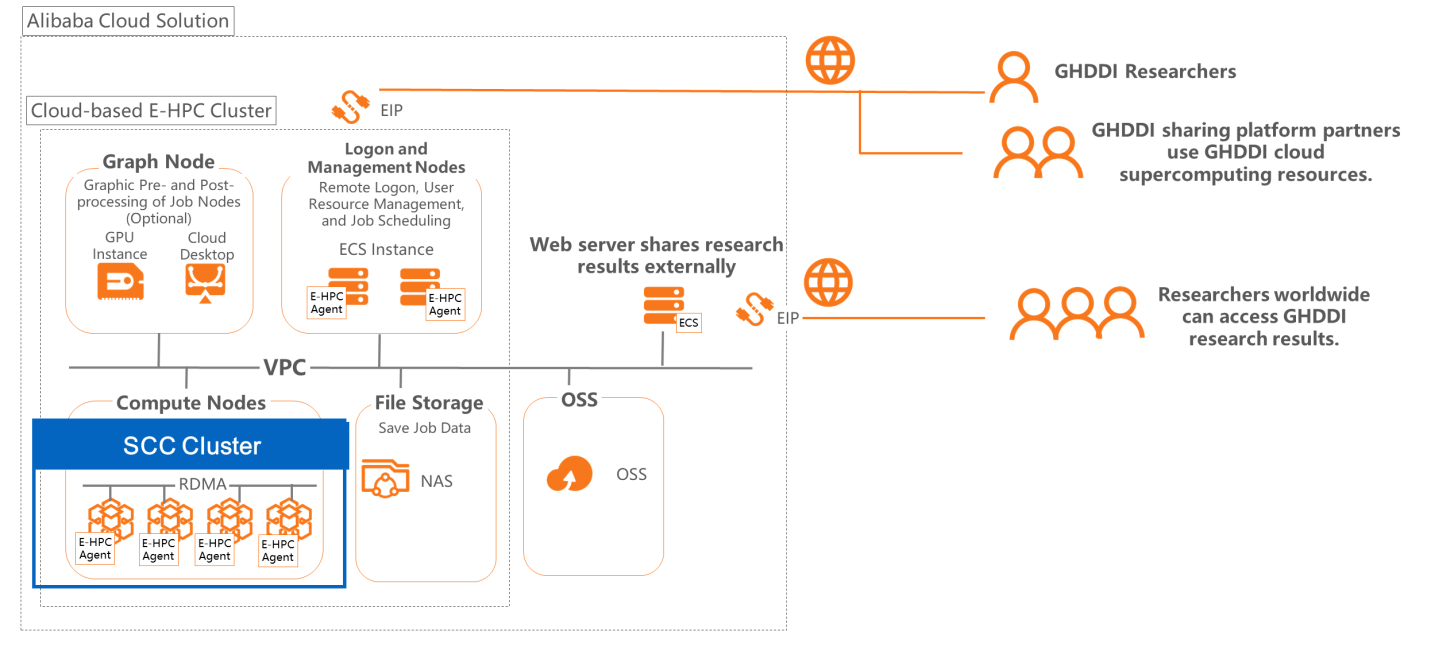
To speed up drug research and development, high-performance computing clusters can serve as an important tool as they can provide powerful computing capabilities to speed up the research process. Researchers can use Alibaba Cloud's E-HPC cloud supercomputing product to quickly build high-performance clusters in the cloud, giving them access to high-performance computing instances that can meet their computing power requirements.
At the same time, E-HPC provides a high-throughput task processing solution that enables drug candidate screening to be concurrently performed on multiple computing nodes and with multiple CPU cores, reducing the overall task execution time. In addition, E-HPC is a cloud-native supercomputing product. So, it can be integrated with other cloud products, such as OSS, allowing you to quickly and conveniently build computing and information publication platforms .
While continuing to wage war against the worldwide outbreak, Alibaba Cloud will play its part and will do all it can to help others in their battles with the coronavirus. Learn how we can support your business continuity at https://www.alibabacloud.com/campaign/supports-your-business-anytime
How Alipay Has Been Providing 24/7 Services since Spring Festival
How PolarDB Produced Scores for 1.2 Million Online Test Takers in 10 Minutes

2,599 posts | 764 followers
FollowMichael Njunge - November 29, 2022
Alibaba Clouder - April 2, 2020
Iain Ferguson - April 18, 2022
Alibaba Clouder - April 29, 2020
Alibaba Cloud ECS - March 5, 2021
Alibaba Clouder - April 15, 2020

2,599 posts | 764 followers
Follow Platform For AI
Platform For AI
A platform that provides enterprise-level data modeling services based on machine learning algorithms to quickly meet your needs for data-driven operations.
Learn More Container Service for Kubernetes
Container Service for Kubernetes
Alibaba Cloud Container Service for Kubernetes is a fully managed cloud container management service that supports native Kubernetes and integrates with other Alibaba Cloud products.
Learn More MaxCompute
MaxCompute
Conduct large-scale data warehousing with MaxCompute
Learn MoreMore Posts by Alibaba Clouder