Overview
On February 5, 2020, the National Health and Medical Commission of China issued Pneumonitis Diagnosis and Treatment Program for New Coronavirus Infection (Trial Fifth and Later Edition), which highlighted that the recommended medical evidence for the diagnosis of pathogens are real-time fluorescent RT-PCR of respiratory specimens or blood specimens for detection of novel coronavirus nucleic acid; and sequencing of viral genes in respiratory specimens or blood specimens, highly homologous to known new coronaviruses.
The technology for whole genome sequence analysis is a technology for virus genome sequencing from sample to report. It establishes virus screening and analysis capabilities for local disease control centers and customs agencies that need to manage COVID-19. It provides viral genetic data screening, automated analysis and report generation, and can also work as a research platform for laboratories with experimental and sequencing capabilities.
Through optimization of algorithms and models, it can complete the test of the new coronavirus within 14.5 hours with a high throughput of 20 samples (depending on the sample size and sequencing throughput). It helps solve key issues in current testing approaches, such as insufficient nucleic acid detection capacity, high false negative rate of PCR method, and possible mutation of the virus worldwide.
Technology Highlights
-

Rapid and accurate testing to improve the handling of local outbreaks
Based on the whole-genome sequencing method, the relevant detection is supposed to be accurate and effective in preventing ignorance of viral mutations. The protocol integrates the advanced technologies related to experimental libraries, computer sequencing, and data analysis, reducing the time of sequencing methods and increasing laboratory treatment throughput.
-

Core algorithm optimization and comprehensive monitoring of COVID-19 development and changes
This technology analyzes the characteristics of the COVID-19 virus genes; optimizes the training procedure of algorithms based on data from public datasets; designs distributed and parallel algorithms to speed up the analysis process; and develops a rapid virus stitching method. What's more, the design of evolutionary analysis and protein structure analysis based on AI algorithms helps discover the evolutionary source, evolution time, and three-dimensional structure of the virus. The solution also provides a basis for the development of virus vaccines and drugs.
-

Quick deployment and easy to use with modular and simplified operation and configuration
The technology seamlessly connects the library building instrument, sequencer, and computer analysis machine to achieve the complete automatic process, from sample to report. It can be rapidly deployed and provides a web-based operation interface, allowing, front-end personnel to start the analysis of a sample with a single click and directly view the final report. In addition to the complete system, it can also dock customers' existing experimental and sequencing equipment.
Genome Sequencing from Sample to Report
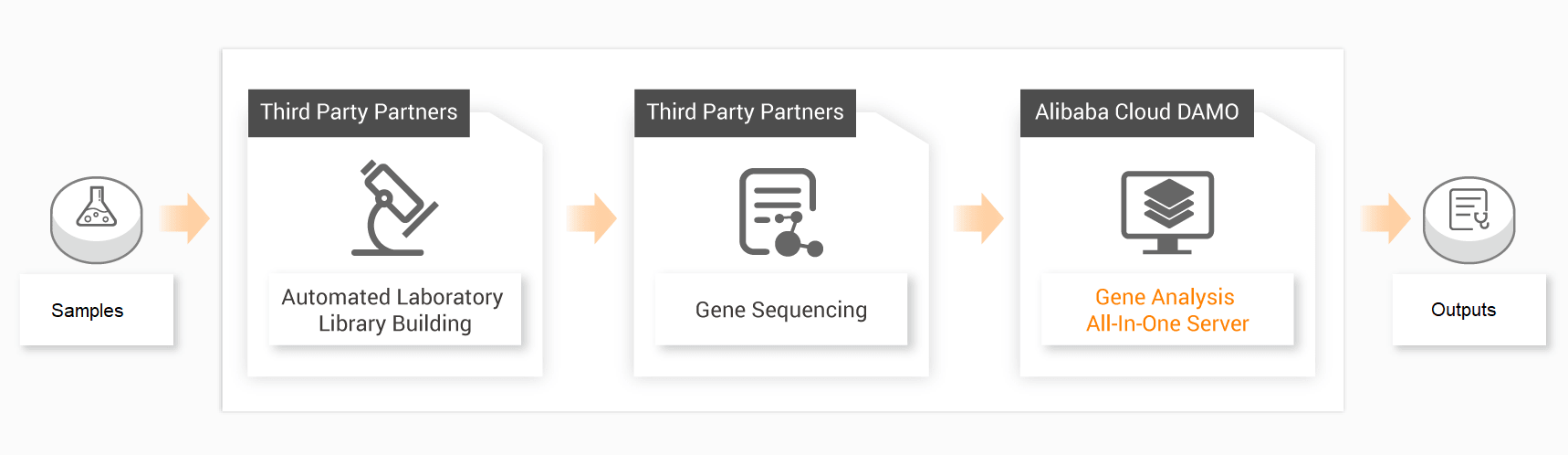
This technology for whole genome sequence analysis generates three primary outputs.
● Homology Analysis Report: Analysis of homology to viral sequences by sequence analysis and sequence splicing to form customized final report.
● Evolutionary Analysis: Evolutionary tree construction, time map for intelligent analysis of viral transmission or evolution, intelligent analysis of the infection time.
● Structure Analysis: Prediction of viral protein secondary structure and three-dimensional structure.
Rapid and Accurate Testing
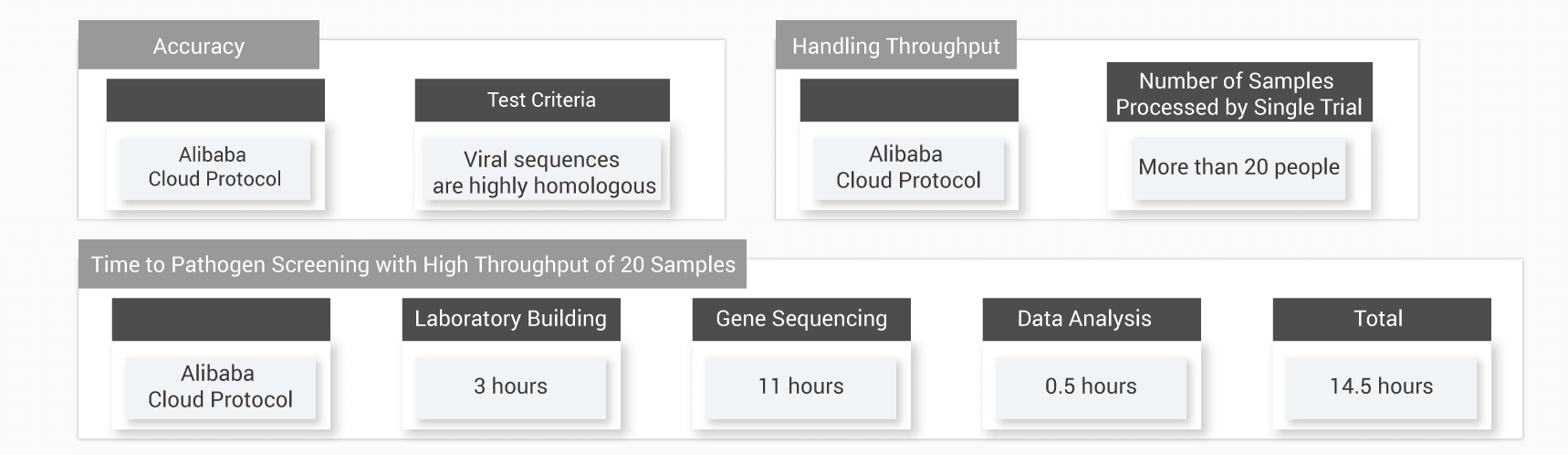
This technology greatly reduces the data analysis time to 0.5 hours for an experiment of 20 samples in parallel, and is able to test one sample within 43.5 minutes.
Note: The time of 43.5 minutes is calculated based on a sample size of 30-100GB tests and on NextSeq 500. The total test time is 14.5 hours for every experiment of 20 samples in parallel, in which 3 hours for laboratory building; 11 hours for gene sequencing; and 0.5 hours for data analysis powered by this technology for whole genome sequence analysis technology. The time of completing one sample test is 43.5 minutes by dividing the 14.5 hours by 20 samples. This test result shows a variable time, depending on sample size and sequencing throughput.
Free for Public Research Institutions
Free Computational and AI Platforms to Help Research, Analyze and Combat COVID-19 ("Program") is intended to support disease control centers and genome research institutions worldwide for the research analysis and prevention of COVID-19. You can submit the summary and description of your research project to wanqing.hwq@alibaba-inc.com. All submissions will be reviewed for technical feasibility, and eligible applicants will be contacted for more details and the next steps of the process. Once the submission is successful, the applicant will get a certain amount of coupon according to the project, which is valid for 3months. The coupon can be used for all Alibaba cloud products, including HPC, ECS, and GPU, excluding marketplace products and 3rd party products.
Apply NowRelated Resources
Whitepaper
Alibaba Cloud Helps Fight COVID-19 through Technology
Highlights Alibaba Cloud's cloud technologies, programs, and initiatives that are helping fight against the threat of COVID-19.
Whitepaper
Whole Genome Sequencing Analysis for COVID-19
Explains technologies that help to accurately detect virus mutations and reduce the genetic analysis time.
Webinar
Alibaba Cloud DAMO Whole Genome Sequencing Analysis
This webinar explains how Alibaba Cloud DAMO Academy use genome technologies to fight COVID-19.

