CRISPR-Cas9 is a genome editing tool that is creating a buzz in the science world. It is faster, cheaper and more accurate than previous techniques for editing the genome of living cells. It hence has the potential to revolutionize a wide range of applications.
CRISPR-Cas9 has a lot of potential especially in the health space as it allows the treatment of medical conditions that have a genetic component, including cancer, hepatitis B or even high cholesterol. Clinical trials have already started for patients with specific blood and solid cancer types.
CRISPR-Cas9 is suitable for these applications because it can be programmed to recognize and edit specific locations in the genome by pattern-matching unique sequences of DNA. However, for robust application in the clinic, the efficiency of CRISPR-Cas9 needs to be increased as does the speed with which target sites can be designed.
Researchers in the eHealth program of the Commonwealth Scientific and Industrial Research Organization (CSIRO) in Australia, developed GT-Scan2, a novel software tool to address both issues.
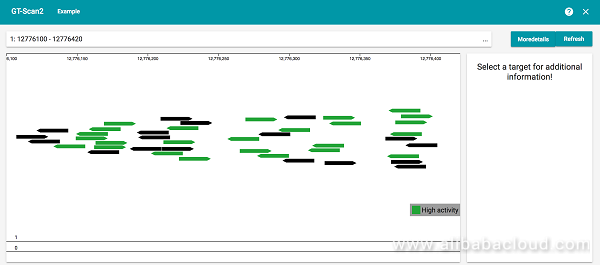
GT-Scan2 can help researchers find the most effective CRISPR/Cas9 targets in a genomic region by ranking targets by the predicted cutting efficiency. You can think of it as the "search-engine for the genome". GT-Scan2 will also report the number of potential off-targets for each target, where potential off-targets are other regions in the genome with 0-3 mismatches to the target.
A Web Application front end is used to access the GT Scan2 application and to submit the relevant jobs.
When a user submits a job, GT-Scan2 inserts the job parameters as an item into a TableStore table via an API call. This allows the solution to be freely scalable without creating a bottleneck. The database entry triggers the first Function Compute function, which finds all putative CRISPR targets in the user-specified DNA sequence (fetched automatically upon user submission). Potential CRISPR target sites have fixed rules and can be easily found using a regular expression that completes in seconds and are inserted into a second TableStore table.
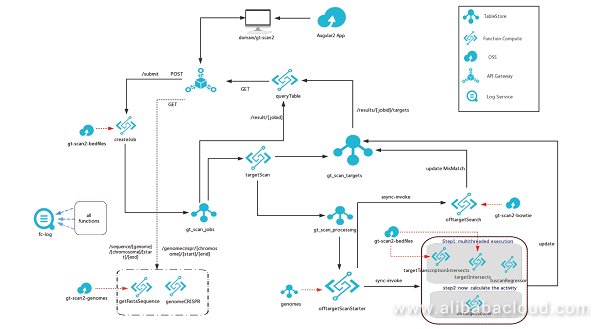
GT-Scan2 is served directly from OSS making it a static web app without server-side processing. It retrieves the dynamic content (such as job results and parameters) via API calls using API Gateway from a NoSQL database (TableStore) using a JavaScript framework.
All potential targets need to be evaluated for their off-target risk using the efficient string matching tool, Bowtie. Though Bowtie only requires a reduced representation of the 3 billion letter genomic sequence, the size of these index files still reaches 915 MB for the human genome. Even though Alibaba Cloud Function Compute supports temp spaces of this size, the implementation divides the genome into smaller blocks to enable parallel processing. For an average run, GT-Scan2 hence triggers 200-500 individual Function Compute functions, which simultaneously update the scores for the different putative targets in TableStore. During this process, the frontend is polling this table via API Gateway and updating the webpage as results come in, eliminating the need for server-side compute.
Alibaba Cloud Function Compute provides a framework to develop a future-ready software package that is able to support medical genome engineering applications. It has the ability to instantaneously scale at run time to the optimal capability by spawning the appropriate number of functions to cope with the varying complexity of different genes. Other benefits include only paying for the storage when no compute is triggered; jobs not competing with web server resources as the website is a static page with dynamic content being updated through Angular 2 and the API Gateway; as well as not needing to maintain compute instances (security patches of OS).
GT-Scan deployment benefitted from the Alibaba Cloud specific architectural patterns and services. Some of them are listed below.
The open sourced Fun Tool (Fun with Serverless) will enable automated deployments of API Gateway and Function Compute resources making deployments of new GT Scan versions a breeze. The tool allows automated deployments of components defined in a simple YAML file.
Leverage Alibaba Cloud's award winning big data platform to create a Machine Learning Pipeline will enable sophisticated analyses to be integrated in the application. This is of specific relevance for personalized health applications, which identify editing strategies for individual patients.
Alibaba Cloud Log Service allows exporting log files for future analysis leveraging Alibaba Cloud's big data platform of existing open sources analysis platforms available at CSIRO's disposal. The log file exports can then be plugged into an existing machine learning pipeline to learn from the usage patterns of the GT-Scan application.
Alibaba Clouder - June 12, 2020
Alibaba Container Service - July 16, 2019
Alibaba Cloud Community - January 20, 2022
Alibaba Clouder - April 26, 2020
Alibaba Cloud Native - August 14, 2024
Alibaba Container Service - July 1, 2024
 Big Data Consulting for Data Technology Solution
Big Data Consulting for Data Technology Solution
Alibaba Cloud provides big data consulting services to help enterprises leverage advanced data technology.
Learn More Big Data Consulting Services for Retail Solution
Big Data Consulting Services for Retail Solution
Alibaba Cloud experts provide retailers with a lightweight and customized big data consulting service to help you assess your big data maturity and plan your big data journey.
Learn More Serverless Workflow
Serverless Workflow
Visualization, O&M-free orchestration, and Coordination of Stateful Application Scenarios
Learn More ApsaraDB for HBase
ApsaraDB for HBase
ApsaraDB for HBase is a NoSQL database engine that is highly optimized and 100% compatible with the community edition of HBase.
Learn MoreMore Posts by Sabith